Denisko D, Viner C, Hoffman MM. Motif elucidation in ChIP-seq datasets with a knockout control. Bioinformatics Advances, 2023. Available from: https://doi.org/10.1093/bioadv/vbad031
PeaKO identifies motifs relevant to ChIP-seq experiments by combining two differential analysis approaches. It often improves elucidation of the target motif over other methods and highlights the benefits of knockout controls.
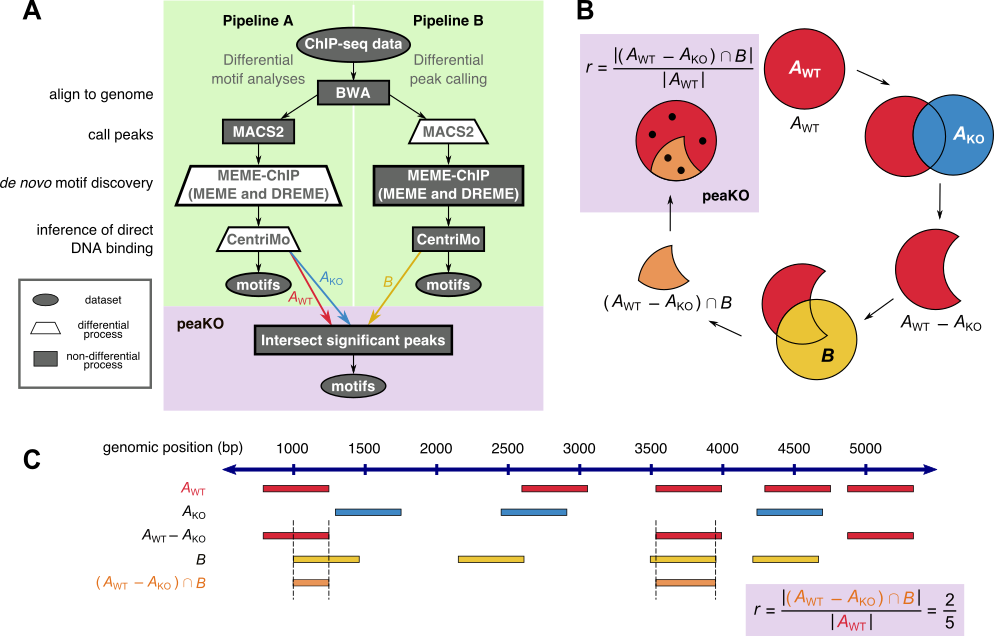
PeaKO is a computational method for identifying motifs from wild-type/knockout paired ChIP-seq datasets. PeaKO optimizes motif analyses by implementing a dual-pipeline approach. The first pipeline incorporates differential motif analysis, while the second incorporates differential peak calling. We combined these pipelines to select for motifs that both have consistent matches within peaks and fall within regions of significant read pileup. PeaKO computes a new metric based on the proportion of overlapping peaks between both pipelines, with overlaps interpreted as genuine binding events. PeaKO uses this metric to rank a collection of known or de novo motifs, where top-ranked motifs are thought to be relevant to the ChIP-seq experiment.

PeaKO can be used in a variety of contexts including motif enrichment and motif discovery. PeaKO automates the entire process starting from aligned reads including both pipelines, so users must only provide wild-type and knockout BAM files along with associated reference genome files and a motif database file. We ran peaKO on 8 publicly available wild-type/knockout paired sequence-specific transcription factor ChIP-seq datasets, including ATF3, ATF4, CHOP, GATA3, MEF2D, OCT4, SRF, and TEAD4.
Poor antibody quality, characterized by low specificity to the target transcription factor or non-specific cross-reactivity, presents a major source of noise in ChIP-seq experiments. Common types of control experiments, such as input and mock immunoprecipitation controls, suffer from a range of issues and fail to account for the full extent of noise. Knockout controls present an attractive alternative to input and mock immunoprecipitation. In these experiments, mutations directed to the gene encoding the target transcription factor result in little to no expression of the transcription factor, prior to ChIP-seq. This preserves most steps of the ChIP protocol, including antibody affinity purification. Therefore, KO experiments can account for both antibody-related noise and biases in library preparation. PeaKO presents an optimal computational processing workflow when knockout controls are available.
pip.
Please read peaKO's documentation for installation instructions.
We have only tested this software on Linux systems and cannot ensure proper installation or functionality on other systems.
It can run either locally or on Slurm cluster systems (using the template configuration file below).
PeaKO was developed by Danielle Denisko, Coby Viner, and Michael Hoffman.